School of Plant Sciences Seminar Series
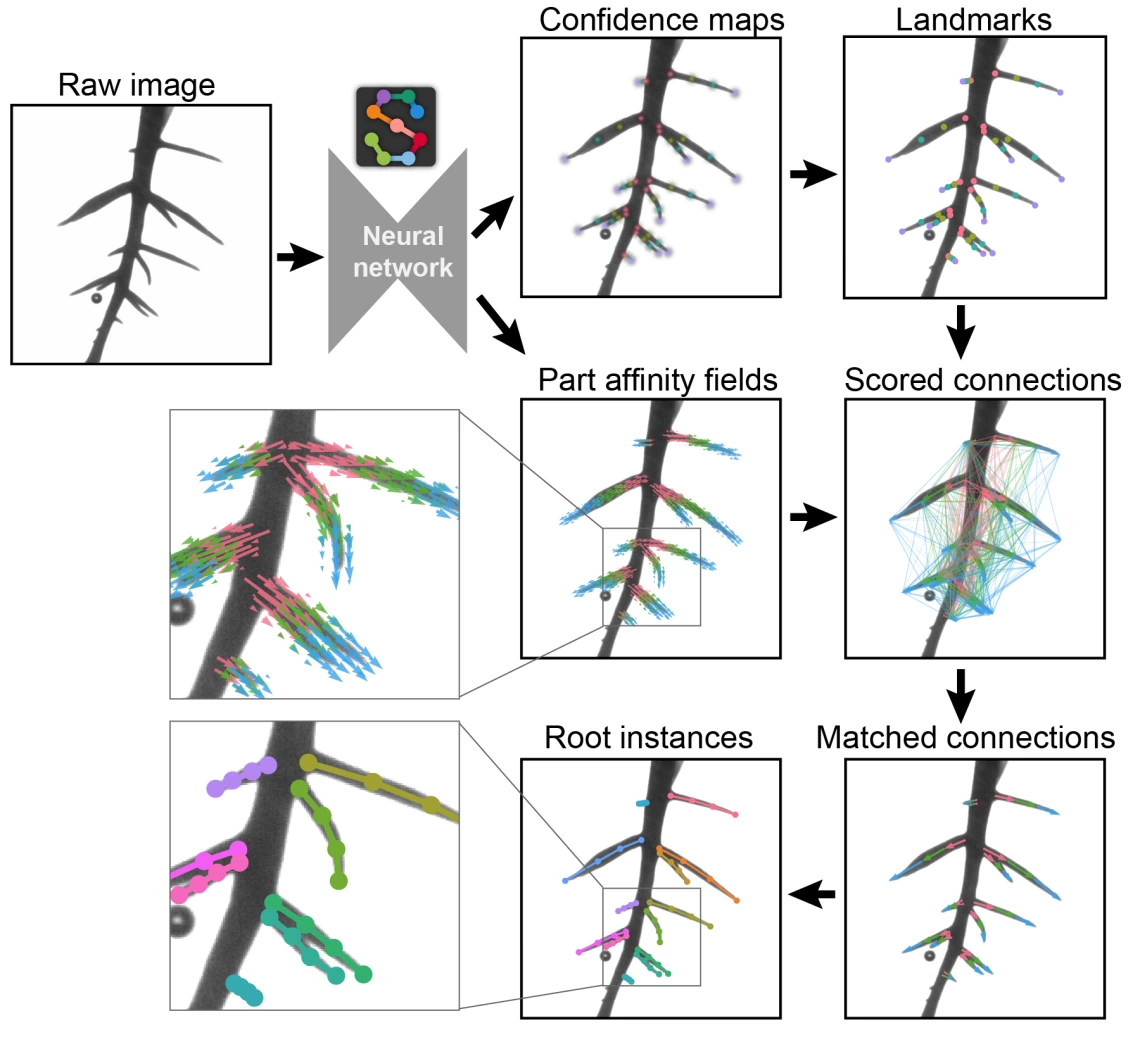
Fig. 1. Demonstration of the bottom-up method for root pose estimation. A detailed image of a root system cultivated in a clear gel cylinder is input into a trained NN via SLEAP. This network concurrently produces confidence maps and part affinity fields. The confidence maps, with distinct color codes for each landmark type (r1 in pink, r2 in yellow, r3 in green, r4 in purple), pinpoint probable root landmark positions. These are then sharpened into discernible peaks based on the highest probabilities within each map. Part affinity fields encode part-to-part associations in 2D vector fields across the image. Using these data, connections between root landmarks are evaluated and scored. The most highly scored connections serve to pair the root landmarks, culminating in the assembly of individual root instances, each distinguished by a unique color.
Elizabeth Berrigan
Speaker
When
Where
Abstract: High-throughput root phenotyping is essential for understanding plant development, stress resilience, and crop improvement. In this talk, I will present an automated, deep learning-based pipeline for root system phenotyping that leverages pose estimation to extract morphological traits with high accuracy and minimal annotation. By integrating machine learning model generalization techniques with scalable data management and MLOps pipelines, we enable reproducible and efficient root trait analysis across diverse species. This approach significantly reduces manual effort, improves model robustness, and facilitates large-scale, data-driven discoveries in plant biology.
Bio: Elizabeth has a master’s in physics from the University of Chicago where she studied the relationship between hydrodynamics and string theory. As an undergraduate, she spent time at MIT studying the time evolution of entanglement entropy using the holographic principle in a system approaching equilibrium to understand how different quantum subsystems are correlated over time with Professor Hong Liu. After her Master’s, she went home to New Hampshire and worked on a farm and at a community art center that sold sustainably made crafts. She is excited to be in the Busch Lab, using her quantitative skills to serve HPI’s goals. She is interested in developing deep learning pipelines for image analysis of roots in crops and using machine learning techniques for data analysis. She is at the greenhouse in Encinitas working alongside the phenotyping team as a bioinformatics analyst I. When she isn’t phenotyping roots, Elizabeth is tutoring math and physics or enjoying sustainable crafts such as wood-burning and making botanical dyes. She loves the outdoors.

